Swarbreck Group
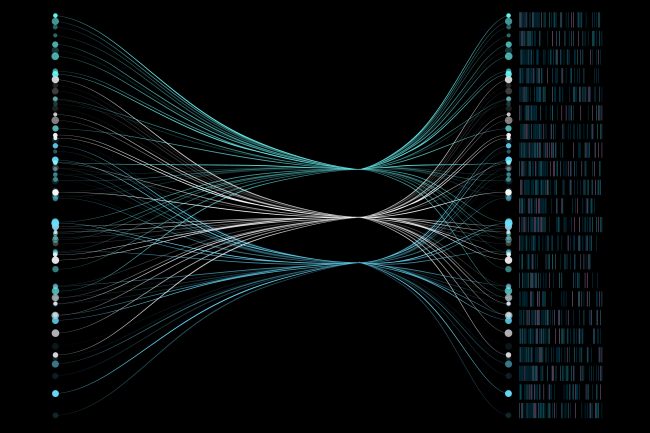
Understanding the complexity of eukaryotic transcriptomes and gene regulation
Understanding the complexity of eukaryotic transcriptomes and gene regulation
The Swarbreck Group (Core Bioinformatics) provides high-quality computational analyses of a range of sequence data types and develops software and analysis pipelines that support the Institute’s Strategic Programmes and National Bioscience Research Infrastructure in Transformative Genomics.
The group are involved in the analysis of sequence data from a wide variety of sequencing platforms and work alongside other Earlham Institute research groups to establish new software tools and analysis methods.
We have a specific research interest in the development of sequencing, assembly, and annotation strategies to characterise transcriptome complexity and the application of RNA sequencing coupled with complementary data (Chip-seq, BS-seq, ribosomal profiling) to explore the complexity of eukaryotic transcriptional landscapes.
Mikado
A method to leverage multiple transcriptome assembly methods for improved gene structure annotation.

Portcullis
A tool for junction analysis and spliced read alignment filtering.