Targeting global food insecurity with sustainable wheat
Wheat provides around a fifth of the world’s daily calories and protein. And in some countries, it accounts for half of the calories consumed.
It’s eaten across the world, in everything from bread, cereal, and pasta, to noodles, couscous, and tortillas. Its use in animal feed means even meat is wheat-dependent. Our global population is growing so, to ensure everybody is fed, wheat production needs to increase as well.
But more than half the world’s wheat supply comes from just five countries, one of which – Ukraine – is currently at war.
Any increase in wheat production must avoid equivalent growth in environment-threatening fertilisers, as well as navigating the extreme weather events and new crop diseases of the escalating climate crisis.
With wheat expected to be feeding 10 billion people by 2050, and climate change and war blighting crops, the stakes have never been higher - or the footing more precarious.

The Earlham Institute, an integral part of the international effort that delivered the first complete bread wheat genome in 2014, is a core partner in a collaborative initiative to safeguard wheat for humanity.
The Delivering Sustainable Wheat (DSW) programme, led by the John Innes Centre, is built on decades of previous work on this staple crop to which EI researchers made fundamental contributions.
The programme is strategically funded by the Biotechnology and Biological Sciences Research Council (BBSRC), part of UKRI.
The programme has four strands: Targeted Sustainability-trait Discovery, Delivering Resilience to Biotic Stress, Nutritional Traits, and Sustainable Data Frameworks for Wheat. Earlham Institute researchers are involved in the first and second strand, and the Institute is co-leading on the fourth.
Dr Richard Leggett, Group Leader at the Earlham Institute, is co-leading the Sustainable Data Frameworks for Wheat work package.
This will concentrate on standards, sharing, connectivity, and infrastructure, with a special emphasis on FAIR (findable, accessible, interoperable and reusable) data.
“Open and FAIR data access is an absolutely crucial part of this programme,” says Richard.
“The work done as part of DSW will be used by plant breeders and wheat researchers around the world, potentially for years.
“Being able to share data in a standardised way will be vital for achieving sustainable production of this important crop in the coming decades.”
Earlham Institute-developed systems playing a part in DSW include MARTi, a software package designed for real-time nanopore sequencing and analysis of metagenomic samples.
And Grassroots was created to be a community-led resource to engage wheat researchers - from breeders to bioinformaticians - in generating, evaluating and integrating wheat data.
Over the past two decades, advances in high-throughput genomics have meant a dramatic increase in digital data generation within wheat research.
Delivering assembled and annotated genomes for key strategic cultivars, and accelerating the use of these resources in research and breeding, is a key goal for DSW.
The Earlham Institute’s expertise in data standards, infrastructure, and sharing mean it can develop long-term data strategies for DSW, giving the project sustainable data coordination.
But DSW is not just about safeguarding our wheat. What if we could not only increase the production of wheat… but improve it?

Air samplers set up at the John Innes Centre Field Trials at Church Farm.
The Earlham Institute’s approach for detecting airborne pathogens, AirSeq, will be used to establish a pathogen species baseline for UK wheat. This will be used to explore pathogen population dynamics and the emergence of new threats.
And the Institute, in collaboration with Rothamsted Research, is working on the wheat root pathogen take-all, the most important root disease of wheat worldwide.
Take-all is caused by the fungus Gaeumannomyces tritici. It destroys around a fifth of wheat yield annually in the UK.
“This fungus belongs to an important group of grass-associated fungi, including pathogens causing rice and wheat blast diseases,” says Dr Rowena Hill, a Postdoctoral Scientist working across the Neil Hall and Anthony Hall Groups.
“However, take-all of wheat has been an understudied disease, perhaps due to difficulties associated with identifying and isolating it from roots or soil.
“Little is known about how far the fungus spreads, or its geographical range.”
Dr Mark McMullan is a Population Genomicist in the Neil Hall Group. He is heavily involved in the second strand of DSW.
“We have genome sequenced several isolates of the fungus - and also related pathogen and non-pathogen species - in order to try to understand what it is about take-all that makes it pathogenic,” says Mark.
“We’ve used these genome sequences to generate a UK pangenome, and we are already seeing differences - at both the gene and whole genome level - consistent with increased pathogenicity.”
Mark believes there will be considerable gains for wheat farmers and breeders if the virulence, spread, and pesticide resistance of the fungus is better understood.
DSW is also exploring wheat's potential as a CO2 sink, meaning the vast fields of wheat necessary for our growing population could help us fight the climate crisis. A comprehensive understanding of nitrogen use efficiency and wheat’s adaptation to a changing climate is required.
And nutritional improvements in wheat – more iron, zinc, calcium, and fibre – could help malnourished communities worldwide, reducing healthcare costs and human misery. For the first time, human intervention trials will be conducted to provide direct evidence of physiological benefits of nutritionally-improved wheat.

A wheat stalk affected by the take-all fungus Gaeumannomyces tritici.
Four research institutes – the John Innes Centre (JIC), the Earlham Institute, Rothamsted Research, and Quadram Institute – are joining the National Institute of Agricultural Botany and the universities of Leeds, Nottingham, Lancaster, Bristol, and Imperial College London, to deliver the ambitious objectives of DSW.
To achieve their goals, DSW researchers are using an array of novel technologies to probe the wheat genome, including ultra-long read sequencing and spatial transcriptomics. These experiments are carried out in close collaboration with the Institute’s National Bioscience Research Infrastructures (NBRIs), which provide access to technical expertise and world-class facilities for DSW and other research programmes.
Dr Karim Gharbi, leads the Transformative Genomics NBRI. “We are really pleased to contribute these novel datasets to expand our understanding of wheat and wheat pathogen diversity. Our teams of technical specialists hit the ground running from day one and continually innovate to push the boundaries of wheat genomics," he says.
Consultations and extensive discussions with stakeholders from the wheat community helped shape the programme, making sure it addresses the most important questions for farmers and consumers alike.
The programme invests in pre-breeding, feeding any new traits, genes, knowledge, and new types of wheat more directly into breeding, farming, and food production. This should help any improvements to be swiftly adopted.
Dr Simon Griffiths is Group Leader and Delivering Sustainable Wheat Programme Lead at the John Innes Centre. "This programme brings together the complementary skills of four research institutes and five universities, combined with insights from stakeholders, to contribute toward the JIC vision to support healthy plants, healthy people and a healthy planet,” he says.
"Wheat is a staple crop and analysing older varieties, which have more genetic diversity than modern wheat, allows us to discover genes involved in key yield traits for crop productivity and adaptation.
“We are discovering genes that make wheat more resistant to disease, genes that increase the efficiency of nitrogen use so less fertiliser is needed, and genes that make wheat’s roots grow longer, to improve drought tolerance.
“This knowledge is helping us to develop wheat that is resilient to the challenges of climate change, and will improve worldwide food security and nutrition for generations to come. We have already seen a genetic and genomic technology revolution for human health, and alongside our partners and collaborators - we’re now seeing that revolution in crops."
DSW will be building on information from thousands of wheat genetic lines, decoded as part of its forerunner - the world-leading experimental programme Designing Future Wheat (DFW). The Earlham Institute was also part of this programme.
These lines include genomes from the A.E. Watkins collection of wheat landraces. Held at the John Innes Centre and made up of more than 1,000 lines of bread and pasta wheat, this incredibly rich genetic resource was collected by scientist Arthur Ernest Watkins in the 1920s and 1930s. It holds grains from 32 countries in Europe, Asia, and Africa.
The result is full of geographic and phenotypic diversity, much of which is no longer present in modern commercial wheat varieties. Analysing these genomes is allowing researchers to identify beneficial genes and understand more about their roles.
Prof. Anthony Hall’s group is generating a reference genome for one accession of each of the seven ancestral groups identified in the Watkins collection as well as key strategic UK cultivars.
“The aim is to generate complete genomes, filling missing gaps in our understanding of wheat genetic diversity," he says.
“We also aim to generate the resources in a form that allows researchers and breeders to rapidly integrate this new information in their programmes.”
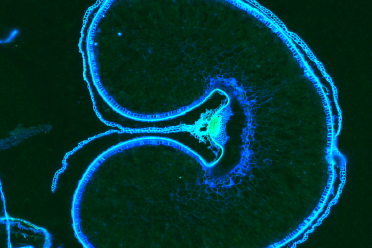
Cross-section of a wheat grain imaged using spatial technologies at the Earlham Institute. ©EarlhamInstitute/Ashleigh Lister
Dr Wilfried Haerty, Group Leader at the Earlham Institute, says the Institute is generating novel genomic resources based on those lines.
“Although all of these wheat varieties were recently sequenced, this was accomplished using short read sequencing and a single reference genome,” he says.
This means that a substantial proportion of the existing genetic diversity inherent to this incredible resource has been either missed or miscalled.”
His group is using novel technologies, including spatial transcriptomics and low input sequencing, to capture genomic regions that would have previously been missed because of their complexity or the use of a single genome.
“This work will provide the necessary baseline to map the unique genomic diversity of this incredible collection and associate this to traits of interest, such as resistance to pathogens, resilience to environmental challenges, and nutritional value,” he explains.
Then both gene editing and traditional breeding could be used to improve crop characteristics - increasing productivity without increasing fertiliser, minimising CO2 emissions, and enhancing wheat's resilience to temperature changes.
From 2012 to 2017, researchers at the Institute worked on the Wheat Institute Strategic Programme (WISP), also contributing to the sLOLA Triticeae Genomics for Sustainable Agriculture grant from 2012 to 2018. And from 2017 to 2023, the Institute collaborated on DFW.
DSW represents a unique, coordinated UK effort to address global challenges in wheat production. Its outcomes could be transformative in addressing the health and hunger of future generations.