A method for assembling the genome of a non-model organism
A walk-through of a manual curation technique for genome assembly
This article is part of our technical series, designed to provide the bioscience community with in-depth knowledge and insight from experts working at the Earlham Institute.
Camilla is a Postdoctoral Researcher in the De Vega Group, specialising in bioinformatics. She is currently working on the genome assemblies for Urochloa decumbens, a tropical forage, and Trifolium pratense, red clover.
The aim is to use the latest technology to generate chromosome-level genome assemblies that can be used to construct pangenomes.
Are you assembling a genome for a polyploid? Do you want to go beyond the traditional collapsed genome and retain all your haplotypes?
This walkthrough is designed to help those interested in using the latest technology, HiFi and Omni-C reads, to generate a chromosome level and haplotype-aware assembly of their own polyploid organism.
This technique can be used for a model or non-model organism. I used it to assemble the genome of signal grass (Urochloa decumbens), a tropical forage grass with complex allelic diversity.
Historically, most genomes that were produced were collapsed representations – a picture of both strands of DNA, combined into one (see Figure 1). This meant, for a heterozygous diploid organism, the assembly would only contain half the information and would not be sorted by haplotypes.
At that time, haplotype-aware assemblies were only possible by sequencing families through trio-binning (short reads generated from two parental genomes are used to sort reads in an offspring’s genome during assembly) or extensive sequencing with multiple technologies.
These strategies were often impractical or prohibitively expensive - but for many analyses a collapsed assembly was all that was needed.
However, recent advances in sequencing technology and a reduction in sequencing costs mean we now have the opportunity to generate haplotype-aware assemblies for non-model organisms.

Figure 1. Diagram showing the two distinct haplotypes present in a diploid genome (A) and how these are represented after being collapsed by a genome assembler.
For orphan crops – my area of work - this represents an invaluable resource for improved breeding.
A large part of this pipeline is adapted from the pipeline developed by the Genome Reference Informatics Team (GRIT) at the Sanger Institute and used by the Darwin Tree of Life Project, on which the Earlham Institute is a partner.
I was fortunate to be trained by the GRIT team, who shared their knowledge about this process and taught me how to identify some common patterns.
The high quality data I used for this project was produced by Tom Barker, Vanda Knitlhoffer, Naomi Irish, Alex Durrant and Fiona Fraser from the Earlham Institute’s Technical Genomics team, and funded by the Biotechnology and Biological Sciences Research Council (BBSRC).
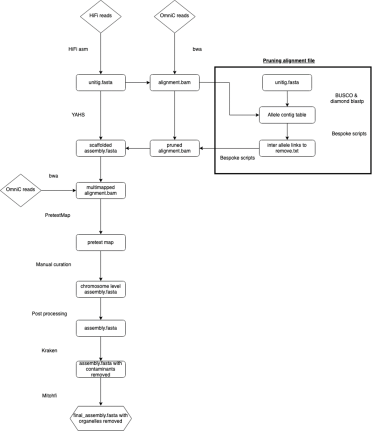
Step one is to build a genome assembly using PacBio HiFi reads and HiFiasm.
Hifiasm produces multiple assemblies at increasing levels of contiguity. Typically, users would take the final phased assembly or the primary contig assembly produced by HiFiasm. However, I decided to use one of the first assemblies produced directly from the reads – the unitig assembly.
There were multiple reasons:

Figure 2. Showing how reads are assembled to unitigs, which are then assembled into contigs. Each step increases the contiguity, but only reads and unitigs are haplotype specific.
The OmniC reads for use in scaffolding were processed using a method called ‘pruning’, adapted from Zhang, et al (2018, 2019). OmniC reads can help a scaffolder understand which unitigs should be placed together, given that sequences close in 3D space should also be close in 2D space.
But while this rule holds true most of the time, this is biology. Sequences may also be close by chance.
Pruning is a way of removing any unhelpful links (Figure 3) existing between sequences close in the 3D space of the nucleus but not close in 2D space.
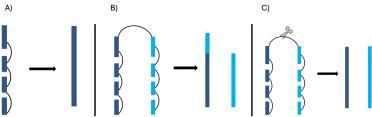
Figure 3. OmniC helps scaffolders by identifying unitigs that are linked (A). however sometimes there are links between unitigs that may be close in the 3D space of the nucleus but are actually from different locations in the genome - these links can confuse the scaffolder and lead to scaffolding errors (B). Pruning (C) removes these unhelpful links.
Firstly, OmniC reads are mapped to the unitig assembly (excluding multi mapping reads). To identify and prune unhelpful links, we created an allele contig table (or, in our case, allele unitig table) by using BUSCO to search the unitig assembly for single copy orthologous genes.
These genes should exist as a single copy per haplotype. If we see them occurring on four unitigs, we can conclude these are separate haplotypes and should not be assembled together. We can therefore remove any links between those four alleles.
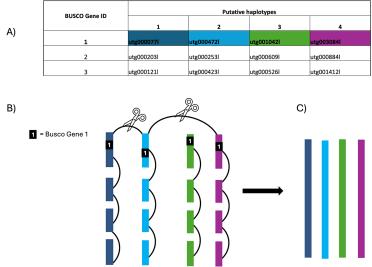
Figure 4. The unitig contig table (A) shows BUSCO genes that have been identified on four unitigs demonstrating they are separate haplotypes. In the above example, gene 1 has been identified on four haplotypes therefore any links between any of these four haplotypes are removed (B) allowing the scaffolder to correctly scaffold four distinct haplotypes (C).
The pruned bam file and unitig assembly are then used as an input for the scaffolder YaHs, generating a more contiguous, scaffolded assembly.
OmniC reads are then mapped to this scaffolded assembly, allowing multi-mapped reads, and the data are processed to generate files for manual curation.
Allowing multi mapping does not help during scaffolding as it creates conflicts. However, during manual curation, it allows you to see certain patterns, such as repeats mapping to multiple locations.
In my opinion, one of the most powerful things about using chromosomal conformation capture technology like OmniC is the ability to visualise and manually curate your genome.
By this, I mean manually identifying assembly and scaffolding errors, correcting them, and further ordering the genome.
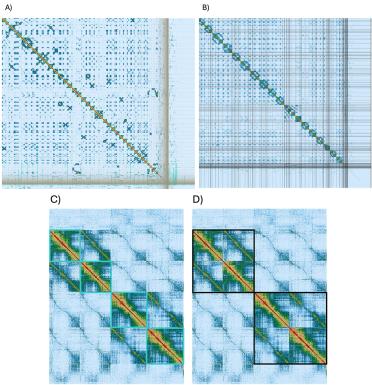
Figure 5. PreText viewer images of (A) the genome assembly before and (B) after manual curation. Division by chromosome is clear (C – outlined in turquoise) as is the division but sub genome (D – outlined in black).
Finally, organelle and contaminant scaffolds and unitigs are identified and removed from the assembly.
Kraken2 and the “Standard-16 nucleotide database” version 2.0.7_refseq-201910 were used to identify any scaffolds that did not belong to Viridiplantae.
Mitohifi was used to identify scaffolds belonging to mitochondria and chloroplast. Mitohifi uses the most similar sequences available (it has a script to determine this) to assist in assembling the two organelles.
For U. decumbens, the most similar Mitochondrial DNA sequence came from Microstegium vimineum (NC_072666.1) and the most similar Cloroplast sequence came from U. decumbens itself (NC_030066.1).
Scaffolds identified as non-Viridiplantae, chloroplast, or mitochondrial were removed from the final assembly.
The final assembly contained 36 chromosomes and 7,086 unplaced scaffolds. This feels like a lot of scaffolds but 85.9 per cent of the assembly content can be found in the 36 chromosomes - including 99.2 per cent complete BUSCO genes from the poales database.
This is very close to the standard set by the Tree of Life project (ToL) for their assemblies (>90% sequence assigned to chromosomes, >90% BUSCO completeness).
However, this assembly is not collapsed and represents every haplotype.
Using this assembly has allowed us to understand the ancestry of polyploid Urochloa decumbens, which has been the subject of discussion within the community for some time.
I hope this assembly will help researchers and agronomists to locate useful genes – such as those responsible for apomixis (asexual reproduction) and pest resistance.
Hopefully this piece will also provide a new approach for you to adapt to your own genome assembly needs.
If you have any questions or feedback - whether positive or negative - please email communications@earlham.ac.uk.