There is a run cartridge that contains large volumes of liquid solutions. This needs to be defrosted an hour before the run. I set this going as soon as possible.
I clear the computer drives of previous runs to ensure we have enough free space to create new raw files. I then clean everything, first with ethanol, then RNAse ZAP spray. I also give the stage on the instrument and microscope a clean.
The instrument requires a wash cycle. This is a good chance for the communications to be tested, and the fluidics too. We run nuclease-free water through to ensure that there are no enzymes in the system that will destroy the RNA or probes during the run.
I check the levels of autofluorescence are acceptable. At this stage, I can top them up before I run the samples.
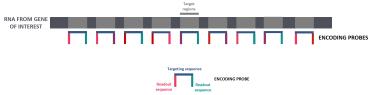
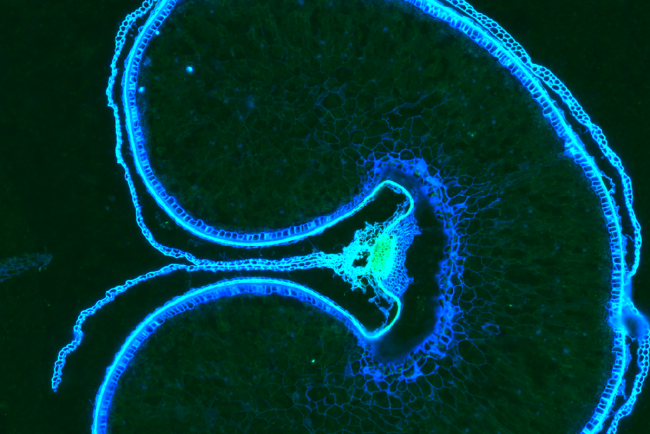
The sample itself is rinsed to stop further digestion. The tissue is stained (different to the probes). The stains are DAPI (which indicates DNA regions in the nuclei of the cells) and poly T (which marks the areas where genes are found).
These are required later on for a processing stage called cell segmentation, when boundaries for individual cells are defined. The stain and the probes are then fixed with more formamide, rinsed, and then the run can be set up.
The slide is fitted into an adapter called the flow chamber. This is a metallic jacket that holds the slide in alignment over the microscope objectives. The other job it does is to create a space that can be filled with fluids. Aqueducts (tubes) deliver various fluids into the flow chamber, from the gene imaging cartridge.
The gene imaging cartridges deliver readout probes - oligos that will hybridise to the handles on the hybridisation probe. There are three that are read out in each round of hybridisation - the number of rounds is different depending on the number of genes you are targeting from your probe panel.
The cartridge also delivers wash fluids to be able to quench (turn off) fluorescence and rinse between readout probe cycles, and also to ensure that the tissue remains wet at all times.
Once the slide is in the flow chamber, seated onto the microscope stage, and the fluidics lines attached, we can trigger the run.
There is an image taken at 10X and the software will show what the DAPI staining looks like at this resolution, as an overview. From this we are able to draw around regions of interest. These are the areas we have told the software to concentrate its imaging efforts on.
Bigger regions mean longer runs and more data, but this is all dependent on the tissue or the research focus.
We can then trigger the 60X imaging. This requires an oil application on the objective. No bubbles must be present. This is the final stage - I can trigger the run, which will take around 12-18 hours to complete. Once the imaging completes, I can set off the processing.
It takes half a day to move the large amount of data (8Tb) to a linux processing computer. The processor stitches all of the z stacks (layers of microscopy images at different thickness depths) at the different locations to make a mosaic as a zipped file.
If we were to run mammalian samples, it’s possible to perform a staining for cell boundaries. If this has been done, the analysis is able to run a python script to segment the cells for you. Plant tissues do not have this luxury and therefore require an off instrument solution, which runs in a similar way.
As a lab technician, my job in the lab is essentially done. All that is left to do is to clean the instrument, and send the data to the HPC using a python script.
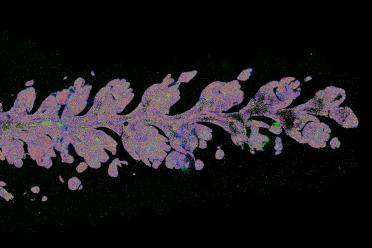
Back in the office, I use software called Vizualiser - a Vizgen-specific image visualisation tool - to look at all of the data as an image. This is the point I know how well it has gone.
I’ll explore all areas of the slide to check the performance of the run. I’m looking for evenness of the staining (the DAPI and polyT), the morphology of the sample (how intacts the tissue is), and the number of spots there are.
Depending on the performance of the run, I will often need to start an email trail, looping in the collaborators to look into troubleshooting with the Field Application Specialist, or their technical support team at Vizgen. They might suggest improvements I could try, or tests I might need to run. This can be put into place for next time.
A batch of slide preparation takes a full week. If this is a big-scale experiment, slides are prepared for running in pairs. One slide is run on Friday, and the other waits until Monday.
Therefore, it might be that once a run finishes on Monday I will set off the prep for the next batches by doing probe hybridisation and in the afternoon set off the run that is waiting.