Yams are a staple part of the Nigerian diet, with Nigeria accounting for around 70% of world yam production, but at current rates of consumption demand is beginning to outstrip supply of this economically important crop with huge cultural significance.
Deciphering the yam genome is of vital importance because, unlike other staple crops such as wheat, maize and rice, the crop is relatively undomesticated. Domesticated crops have advantages compared to their wild relatives when it comes to farming them, including easier usability and higher yields. Understanding the genomics of this crucial plant will help farmers increase yields and sustainability of yams.
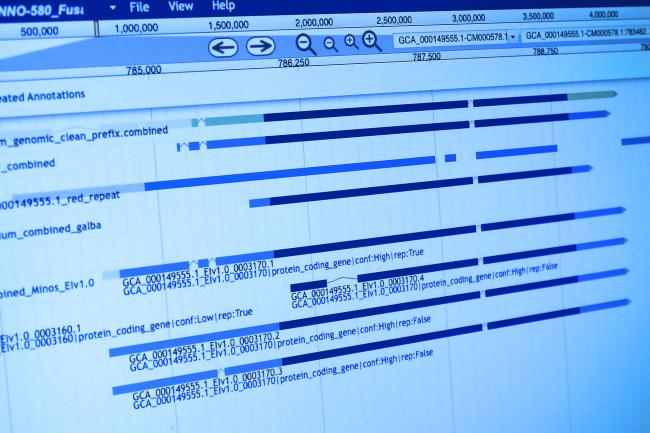
The novel research entitled “Genome sequencing of the staple food crop white Guinea yam enables the development of a molecular marker for sex determination,” is openly accessible in the journal BMC Biology, shows just how that can be made possible. The high quality draft genome sequence is available in the public databases DDBJ and NCBI.
The big breakthrough from an international collaboration of institutes spanning the UK, Germany, Japan and Nigeria, has been in identifying the regions of the genome that determine sex in yams (dioecy), and knowledge of this rare feature is vital for improving the speed of marker-assisted breeding projects.
Dioecy, plants having separate males and females, is relatively rare and occurs only in about 5-6% of flowering plants, including yams and asparagus. Therefore, understanding the process in yams could help in improving other economically important crops.
Most importantly for Central and West Africa, this new knowledge will help transform yams from being a neglected “orphan” crop. With assisted breeding programmes, the crop can be better domesticated, boosting food security and economic well-being in an area undergoing the world’s most rapid population expansion.
Benjamen White, who led Earlham Institute’s contribution, said, “Having a reference sequence for the white Guinea yam gives us the unique opportunity to gain a better understanding of dioecy, a very rare trait in flowering plants, in a species that’s very evolutionarily differentiated from most of what’s been sequenced so far. Understanding this trait and having a genomic resource for white Guinea yam will be invaluable in breeding a better yam, one that will improve food security in West and Central Africa, and the livelihood of smallholder farmers there.”
Dr. Robert Asiedu, Director, Research for Development, for The International Institute of Tropical Agriculture-West Africa, Ibadan, Nigeria, said, “This is an important breakthrough. It means that yam has joined those crops with a full DNA sequence, a development which started with rice some years ago. The implications are profound. The full DNA sequence will greatly facilitate our understanding of the genetic control of key traits such as flowering, diseases, and others including quality traits, and this in turn will make the breeding of new varieties both faster and more precise”.
Professor Ryohei Terauchi, lead author, of Kyoto University and Iwate Biotechnology Research Centre, Japan, said, “This will help to overcome some of the many challenges facing yam farmers in Africa and other parts of the world. These include pests and diseases, post-harvest losses and the need to develop more sustainable systems of farming for the crop”.