Our platforms are strategically supported by BBSRC, part of UKRI, through the Transformative Genomics NBRI.
From isolation through to analysis
We can support all stages of a single- or multi-cell analysis, from cell isolation, to library preparation, sequencing and analysis.
If you are starting with tissue, sorted cells or even pre-made libraries, we have the platforms and expertise to support your project.
Working with us
Whether you are looking for collaboration, contract research or training, our expert team provides access to cutting-edge cell sorting and single-cell genomics infrastructure and expertise.



Our platforms for single-cell isolation
Housed in Biosafety Level 2 Labs, EI has a number of platforms for single-cell isolation for sequencing.
-
-
Spatial Transcriptomics
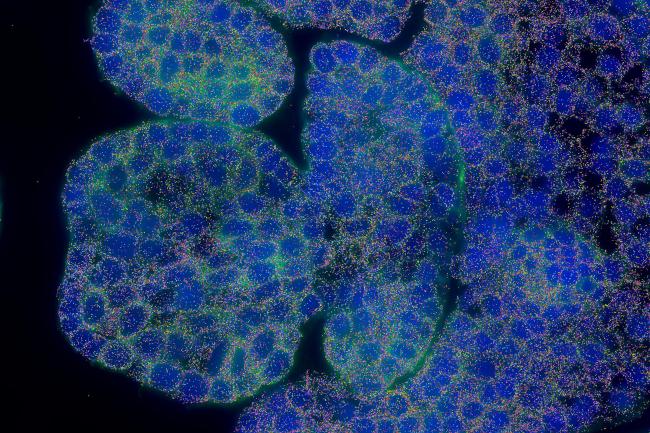
The MERSCOPE™ and MERSCOPE Ultra™ are state-of-the-art spatial genomics platforms that enable the visualization and mapping of hundreds to tens of thousands of RNA species.
The instruments utilise the MERFISH (Multiplexed Error Robust Fluorescence In Situ Hydridization) technology to quantify and map the transcriptome of whole tissues down to a sub cellular level.
The Vizgen MERSCOPE Ultra™ uses the MERFISH technology, with two flow cell sizes - 1.25cm2 and 3cm2 - allowing for larger tissues or more replicates, to fit onto a single slide per run.
The Earlham Institute was an early adopter of this technology. Since the installation of the first Vizgen MERSCOPE platform in 2023, scientists have developed significant expertise in applying these techniques in both mammalian and plant systems, notably collaborating with researchers to spatially resolve 200 genes in developing wheat spikelets.
The Earlham Institute is a Certified Service Provider for the MERSCOPE™ in situ Spatial Transcriptomics Workflow.
-
-
High-throughput single-cell sequencing.
The latest in advanced single-cell analysis technology, the Chromium X platform supports protocols for 3’ and 5’ RNA-sequencing (including V(D)J immune cell profiling) as well as ATAC-seq, CITE-seq and multi-omic profiling.
Chromium X has increased the capacity of the 10X system to enable loading of 16 samples and up to 20,000 cells per sample. The platform also has improved user interface and overall system stability.
-
-
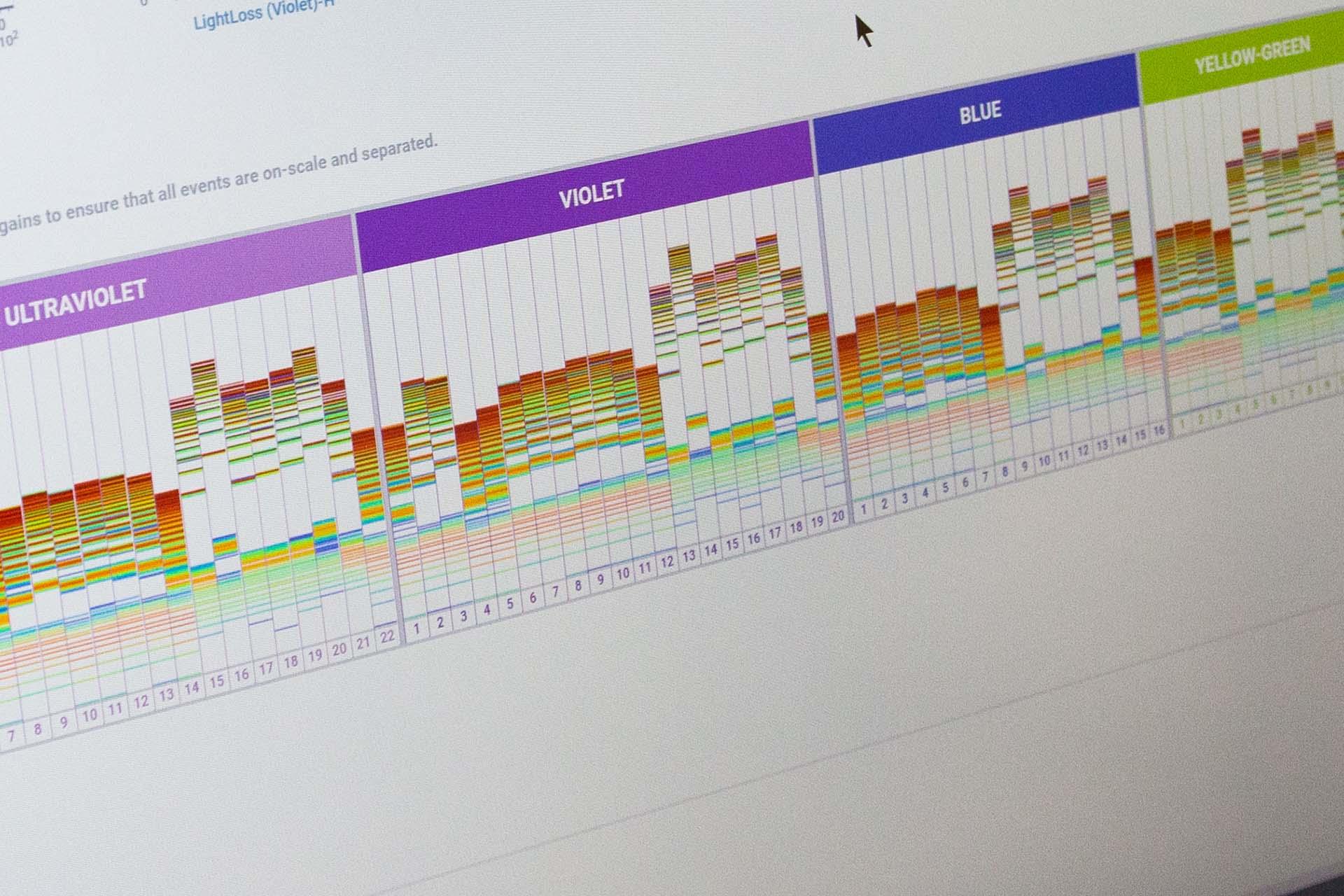
Cutting edge spectral imaging cell sorter.
We are one of the earliest adopters of this groundbreaking platform. Six imaging parameters enable pictures of each cell sorted to be captured in addition to the spectral 78 fluorescence channel detection from 5 lasers. Laser configuration - UV (355nm), Violet (405 nm), Blue (488 nM), Yellow-Green (561nm), Red (640 nm) and index sorting capability.
Imaging parameters captured can now isolate cells based on the morphology of specific cell types. The spectral capabilities facilitate autofluorescence exclusion and multiparameter analysis not previously possible on conventional flow cytometers.
This instrument can sort into 96- and 384- well plates as well as up to 6 population simultaneous sorting.
-
-
Advanced cell sorting enabling high parameter single-cell sorting.
Our advanced cell sorter has a five laser configuration - UV (355nm), Yellow-Green (561nm), Violet (405 nm), Blue (488 nM), Red (640 nm) and index sorting capability.
Enabling up to 20-parameter sorts, the Fusion can be used to isolate extremely rare cells from complex populations, for example, blood stem and progenitor cells or different immune cell populations.
This instrument can sort into 96- and 384- well plates and can also be used for enrichment sorts before single-cell analysis on the 10X Genomics Chromium Instrument.
-
-
Cell sorting and imaging in parallel.
Our cellenONE instrument is similar to FACS, but overcomes some of the challenges encountered when sorting larger, or more fragile cells. Using a glass capillary, individual cells are imaged and gently dispensed into an array of substrates, including 96- and 384- well plates.
The instrument is also equipped with four fluorescence channels (DAPI, GFP, Cy3/RFP and Cy5) enabling live/dead cell selection and isolation of rare cell populations. As the instrument sorts, it images each cell, and so some information about cell phenotype can be associated with any downstream sequencing.
-
-
Isolation of single cells or micro biopsies from tissue sections.
Our laser microdissection platform can image and cut individual cells, or regions of interest, from tissue sections and collect them in tubes or 96-well plates for downstream analysis.
The instrument is a fluorescence microscope, equipped with three fluorescence channels (DAPI, GFP and Cy3/RFP) and high magnification lenses (up to 150X) allowing fine dissection of regions or cells of interest.
Our single-cell sequencing workflows
After cell isolation, we have a number of workflows to generate, sequence and analyse single-cell libraries.

Single-cell library preparation
Our Technical Genomics team offer a complete workflow for Smart-seq2 single-cell RNA-seq analysis, from provision of cell lysis plates through to library preparation, sequencing and preliminary data analysis.
For more bespoke single-cell projects, our single-cell team can assist with cell sorting, 10X Genomics applications and other methods, including single-cell RNA and DNA sequencing on a wide variety of organisms.

Single-cell sequencing
Single-cell sequencing.
Single-cell libraries are sequenced on our Technical Genomics team’s illumina NovaSeq, ideal for cost-effective sequencing of single-cell libraries at any scale.
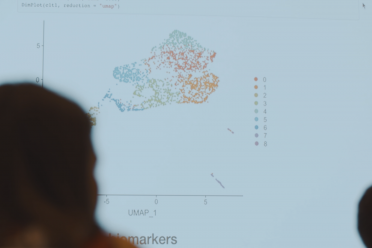
Single-cell data analysis
Single-cell data analysis
EI’s extensive computational infrastructure can support standard analysis of Smart-seq2 or 10X Genomics datasets, data QC, mapping and quantification.
Our in-house expertise can also support more advanced analysis.
Meet the team

Why work with us
Incorporating EI's single-cell genomics in your projects will provide:
- Access to a wide range of platforms to support different sample types - whether it is a standard 10X Genomics RNA-seq experiment, or something more bespoke, we will be able to help
- Experiments run by a dedicated team of experienced single-cell biologists - we have over ten years experience with cell sorting for single-cell and genomics applications, and have published a number of high impact papers and key technical developments in this field
- Additional support and expertise from EI’s Technical Genomics team - from sample receipt to sequencing, all processing is taken care of under one roof. Advanced next-generation sequencing capability and expertise are on hand