Population Variation Genetics
This course will discuss the RAD molecular protocol and the computational analysis of RAD data using Stacks.

This course will discuss the RAD molecular protocol and the computational analysis of RAD data using Stacks.
To familiarise students with the molecular and analytical aspects of the genetic screening method Genotype-By-Sequencing (GBS).
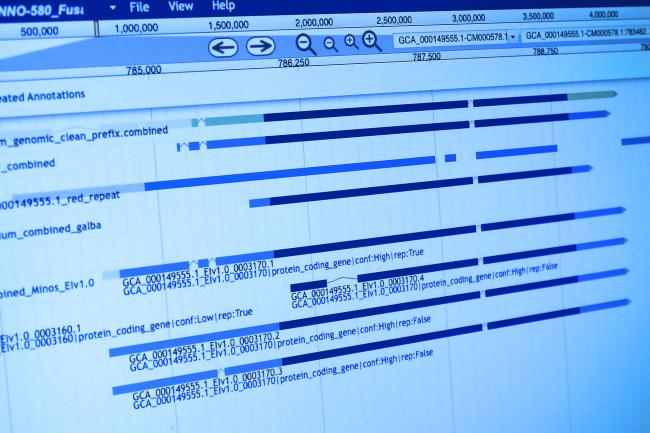
GBS involves the use of short-read sequence data produced from Reduced Representation Libraries (RRL), such as Restricted Site Associated DNA (RAD-tags). GBS libraries can provide a dense set of single nucleotide polymorphisms.
SNP markers are spread evenly across a genome. These markers are useful for a variety of genetic and genomic analyses in both model and non-model organisms. Students will gain experience with the computational pipeline 'Stacks' which was designed for the analysis of such data.
The course will focus on the necessary UNIX skills to handle GBS data, as well as how to clean and demultiplex Illumina sequences. Students will then analyse RAD-seq data de novo to perform a population analysis without the aid of a reference genome using Stacks and structure, and from an organism with a reference genome to generate Fst measures and identify signatures of selection.
Those accepted will be in the planning stages, or producing a Genotype-By-Sequencing project.
Graduate students, post-docs and PI's.
Registration costs are inclusive of lunch and refreshments, course dinner and course materials. Accommodation and transport from/to hotel to EI will be an optional extra during registration to book if you are selected for the course.
Registration deadline: 16 April 2017
Participation: Open application with selection process